2.3.2.26: HECT-type E3 ubiquitin transferase
This is an abbreviated version!
For detailed information about HECT-type E3 ubiquitin transferase, go to the full flat file.
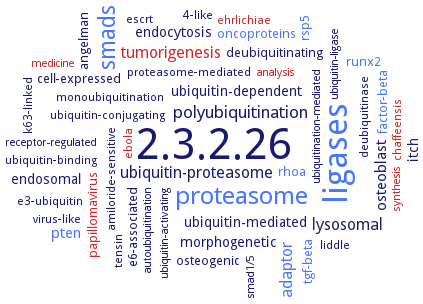
Word Map on EC 2.3.2.26 
-
2.3.2.26
-
ligases
-
proteasome
-
smads
-
polyubiquitination
-
ubiquitin-proteasome
-
adaptor
-
tumorigenesis
-
lysosomal
-
osteoblast
-
pten
-
ubiquitin-dependent
-
endosomal
-
itch
-
ubiquitin-mediated
-
morphogenetic
-
endocytosis
-
factor-beta
-
rhoa
-
osteogenic
-
rsp5
-
deubiquitinating
-
oncoproteins
-
angelman
-
cell-expressed
-
tgf-beta
-
papillomavirus
-
e6-associated
-
runx2
-
tensin
-
deubiquitinase
-
e3-ubiquitin
-
escrt
-
ebola
-
ehrlichiae
-
liddle
-
4-like
-
ubiquitin-binding
-
chaffeensis
-
monoubiquitination
-
virus-like
-
proteasome-mediated
-
amiloride-sensitive
-
k63-linked
-
ubiquitin-conjugating
-
autoubiquitination
-
analysis
-
ubiquitin-activating
-
synthesis
-
ubiquitin-ligase
-
ubiquitination-mediated
-
smad1/5
-
medicine
-
receptor-regulated
- 2.3.2.26
- ligases
- proteasome
- smads
-
polyubiquitination
-
ubiquitin-proteasome
- adaptor
- tumorigenesis
- lysosomal
- osteoblast
- pten
-
ubiquitin-dependent
- endosomal
-
itch
-
ubiquitin-mediated
-
morphogenetic
-
endocytosis
- factor-beta
- rhoa
-
osteogenic
- rsp5
-
deubiquitinating
- oncoproteins
-
angelman
-
cell-expressed
- tgf-beta
- papillomavirus
-
e6-associated
- runx2
-
tensin
-
deubiquitinase
-
e3-ubiquitin
-
escrt
- ebola
- ehrlichiae
-
liddle
-
4-like
-
ubiquitin-binding
- chaffeensis
-
monoubiquitination
-
virus-like
-
proteasome-mediated
-
amiloride-sensitive
-
k63-linked
-
ubiquitin-conjugating
-
autoubiquitination
- analysis
-
ubiquitin-activating
- synthesis
-
ubiquitin-ligase
-
ubiquitination-mediated
-
smad1/5
- medicine
-
receptor-regulated
Reaction
Synonyms
AIP2, AIP4, apoptosis-resistant E3 ligase 1, AREL1, atrophin-1 interacting protein 2, DDB_G0286931, E3 ligase, E3 ubiquitin-protein ligase NEDD4, E3 ubiquitin-protein ligase TOM1, E6-AP, E6AP, ETC-1, HECT domain E3 ubiquitin ligase, HECT ligase, HECT-domain ubiquitin ligase, HECT-type E3 ligase, HECT-type ubiquitin E3 ligase, HECTD3, HectPH1, HECTWWP2, Herc4, Huwe1, Itch, Nedd4, Nedd4 HECT, Nedd4-1, Nedd4L, NEDL1, NleL, RSP5, Smurf1, Smurf2, sog-1, TRIP12, Trp120, UBE3B, Ube3C, ubiquitin-protein ligase E3C, UBR E3 ubiquitin ligase homolog, Ubr-5, UBR1, UBR5, UPL3, UPL5, WW domain-containing E3 Ub�protein ligase 2, WWP1, WWP2

 results (
results ( results (
results ( top
top






